Results
- Target positions.
- Target sequences, 20mer + 3mer PAM (total 23mer).
- GC content of the target 20mer.
- Calculated Tm of the target 20mer.
- Presence or absence of TTTT (four consecutive T’s that cause pol III termination)
in the target 20mer. Avoid TTTT in gRNA vectors with pol III promoter.
- Off-target search results against genomic sequence. The number of target sites with perfect match
is shown. Note that the number displayed here includes both on-target and off-target sites.
Smaller number (but not zero) is better for these columns to avoid off-target editing.
- 20mer+PAM: The target sequence (20mer) adjacent to the PAM is searched against the genome.
One ('1') in this column usually indicates that the sequence has only one perfect match
with the intended target.
Zero ('0') in this column means that the sequence has no match in the genomic sequence;
such sequences may possibly span over exon-exon junctions, so their use should be avoided.
- 12mer+PAM: The 12mer of 3′ region of the target sequence adjacent to the PAM is searched
against the genome. The region contains critical residues determining target specificity.
- 8mer+PAM: The 8mer of 3′ region of the target sequence adjacent to the PAM is searched
against the genome. The region contains the most critical residues determining target specificity.
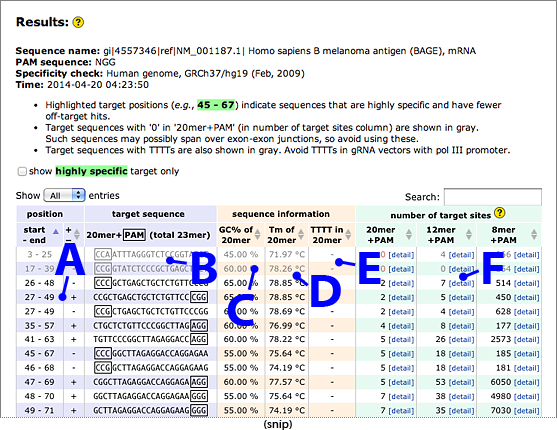
- Only one match in 20mer+PAM and 12mer+PAM search. These targets are highly specific.
→ Recommended for CRISPR/Cas target.
Target positions are highlighted with green (e.g., 163 - 185).
- No match in 20mer+PAM search. Possibly the sequence spans over exon-exon junction,
so avoid using these.
→ Not recommended for CRISPR/Cas target.
- Very high number of off-target hits. Avoid using these sequences.
→ Not recommended for CRISPR/Cas target.


